
For asymmetric logistic curves, a five parameter logistic (5PL) curve is required.įor technical assistance on using this calculator, please contact tool has recently been updated. Note that four parameter logistic (4PL) curves are symmetric in nature around the inflection point.

FOUR PARAMETER LOGISTIC REGRESSION WITH POLYMATH SOFTWARE HOW TO
And it also covers how to do inverse prediction and calculating derived statistics like determining the limit of detection, limit of quantification and working range. STATLIA MATRIX uses powerful numeric algorithms plus accurate weighting with the test’s historical data to provide the gold standard for 5PL curve fitting in analysis software. They also show how one could use weights and iteratively refitted least squares to allow for non-homogeneous variance. Geom_point(data=ame(x=x,y=y), aes(x=x, y=y, ymin=NULL, ymax=NULL), size=5, col="blue") + Geom_line(data=confintervals, aes(x=x, y=fit), colour=I("blue"), lwd=2) + Geom_ribbon(data=confintervals, aes(x=x, ymin=lwr, ymax=upr), fill=I("blue"), alpha=I(0.2)) + Qplot(data=predintervals, x=x, y=fit, ymin=lwr, ymax=upr, geom="ribbon", fill=I("red"), alpha=I(0.2)) + Predintervals = ame(x=xvals,predFit(nlslmfit, newdata=ame(x=xvals), interval="prediction"))Ĭonfintervals = ame(x=xvals,predFit(nlslmfit, newdata=ame(x=xvals), interval="confidence")) They just use the nlsLM function in the minpack.lm package.
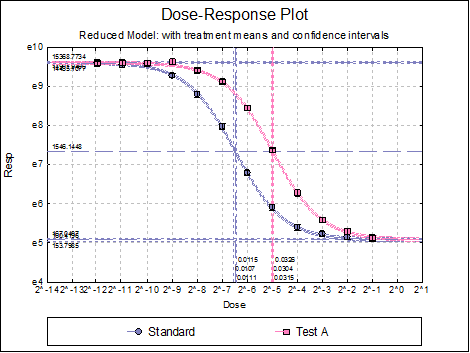
There is an excellent R tutorial on fitting the 4 parameter logistic model for calibration purposes (e.g. Response<-0.5 #lets use 0.5 for the responseĭOSEx<-ED(model1,response,type="absolute",display=F) # the estimated DOSE # type="absolute" gives you the ability to use absolute values for the response, to

# the index (display=F is a good option also) # The result is a matrix, from which the Estimate values can be extracted using # the ED function is used to give the EDx value. If you want to estimate the DOSE from SLOPE, or 'Concentration' from 'OD' in case of an ELISA, just use the ED function of the 'drc' package 'predict' is not the best way to estimate the DOSE from SLOPE in this case, because you have to reverse them in your model2, which doesn't work in this example. "Upper", "ED50")),data=spinach) predict(model2, newdata, Model2 <- drm(DOSE~SLOPE, CURVE, fct=LL.4(names=c("Slope", "Lower", Library(drc) model1 <- drm(SLOPE~DOSE, CURVE,įct=LL.4(names=c("Slope", "Lower", "Upper", "ED50")),data=spinach)


 0 kommentar(er)
0 kommentar(er)
